Generate synthos data
synthos.RmdThe first version of synthos generates semi-realistic
data of three benthic communities: hard coral, soft coral and
macroalgae.
This vignette illustrates an example of how to use
synthos by simulating observations of benthic communities
using the following configuration:
| Main configuration | Example settings |
|---|---|
| Observation data type | Point-based observations |
| Site allocation | Random locations |
| Number of sampling events | 15 years |
| Sampling design | 25 reefs, 3 sites per reef, 5 transects per site, 100 photo frames per transect, 50 points per frame, 2 depths |
| Relative disturbance weights | 80% heat stress, 19% cyclone exposure, 1% other |
| Annual growth values | 3% for hard coral and 3% for soft coral* |
The configuration used in this example can be easily adjusted using the function generateSettings.
*Macroalgae responds differently: instead of growing independently, it occupies the remaining available space (i.e., macroalgae cover = total available space − (hard coral cover + soft coral cover) with total available space is fixed at 80% for each location).
Further explanations of each step can be found in the accompanying vignettes.
1. Generate settings
surveys <- "random" # or "fixed"
data_type <- "points" # or "cover"
synthos::generateSettings(nreefs = 25, nsites = 3, nyears = 15, dhw_eff = 0.5, cyc_eff = 0.3, other_eff = 0.2)2. Generate the spatio-temporal domain, disturbance effects and baselines
spatial_domain <- st_geometry(
st_multipoint(
x = rbind(
c(0, -11),
c(3,-11),
c(6,-14),
c(1,-15),
c(2,-12),
c(0,-11)
)
)
) |>
st_set_crs(config_sp$crs) |>
st_cast("POLYGON")
## ---- SpatialPoints
set.seed(config_sp$seed)
spatial_grid <- spatial_domain |>
st_set_crs(NA) |>
st_sample(size = 10000, type = "regular") |>
st_set_crs(config_sp$crs)
sf_use_s2(FALSE)
benthos_reefs_pts <- synthos::create_synthetic_reef_landscape(spatial_grid, config_sp)3. Generate sampling design
if (surveys == "fixed") {
locs_sf <- synthos::sampling_design_large_scale_fixed(
benthos_reefs_pts, config_lrge
)
obs <- synthos::sampling_design_fine_scale_fixed(
locs_sf, config_fine
)
} else if (surveys == "random") {
locs_sf <- synthos::sampling_design_large_scale_random(
benthos_reefs_pts, config_lrge
)
obs <- synthos::sampling_design_fine_scale_random(
locs_sf, config_fine
)
} else {
stop("surveys must be 'fixed' or 'random'.")
}4. Generate export data table
if (data_type == "points") {
pts <- synthos::sampling_design_fine_scale_points(obs, config_pt)
synthos_data <- synthos::prepare_table(pts)
} else if (data_type == "cover") {
cov <- synthos::sampling_design_fine_scale_cover(obs, config_pt)
synthos_data <- synthos::prepare_table(cov)
} else {
stop("data_type must be 'points' or 'cover'.")
}5. Vizualisations
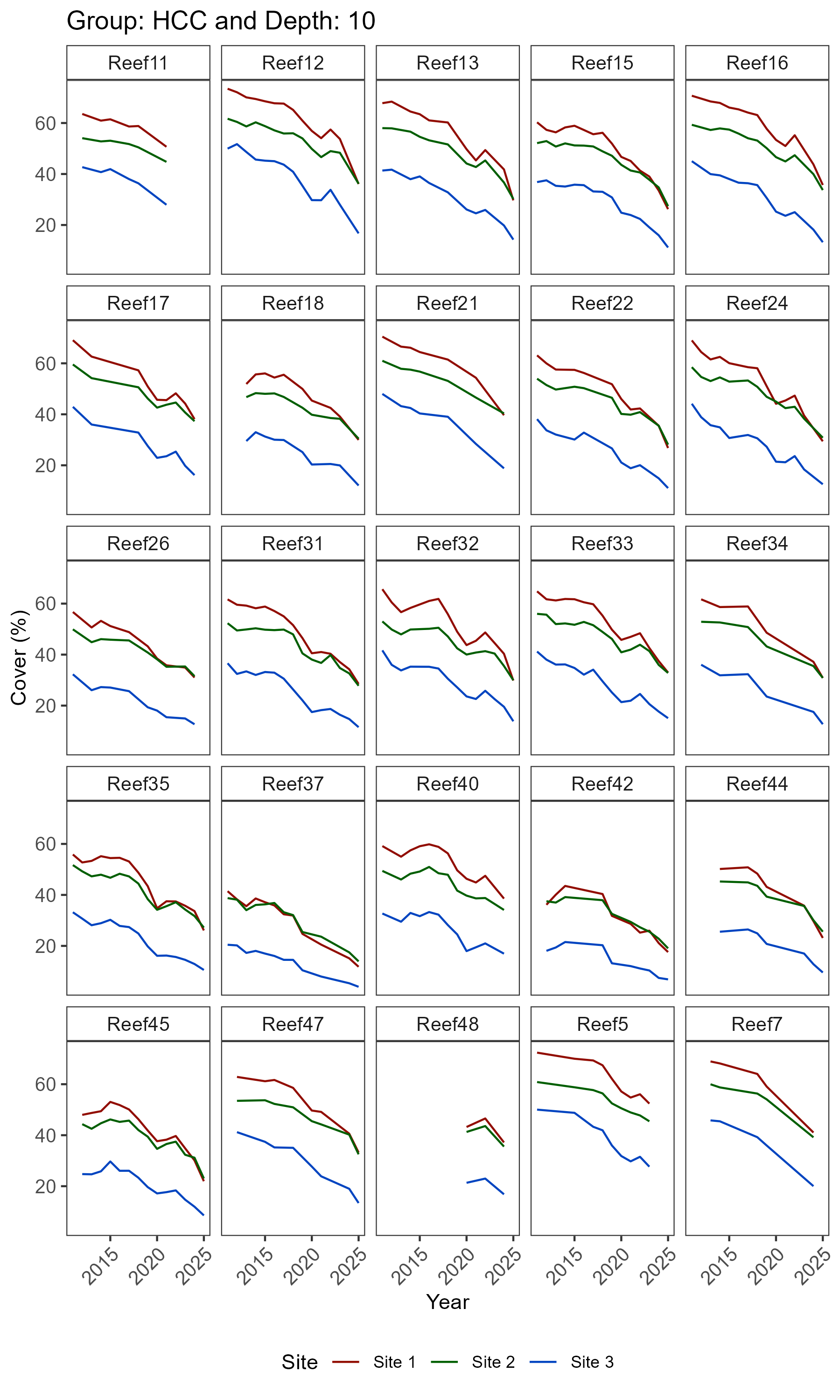
# 3.1 Long-term trajectories at site level
plots_1 <- synthos::plot_synthos(synthos_data, type = "trajectories")
purrr::walk(seq_along(plots), ~ ggsave(filename = paste0("figures/figure1.", .x, ".png"),
plot = plots[[.x]], width = 6, height = 10, dpi = 300))
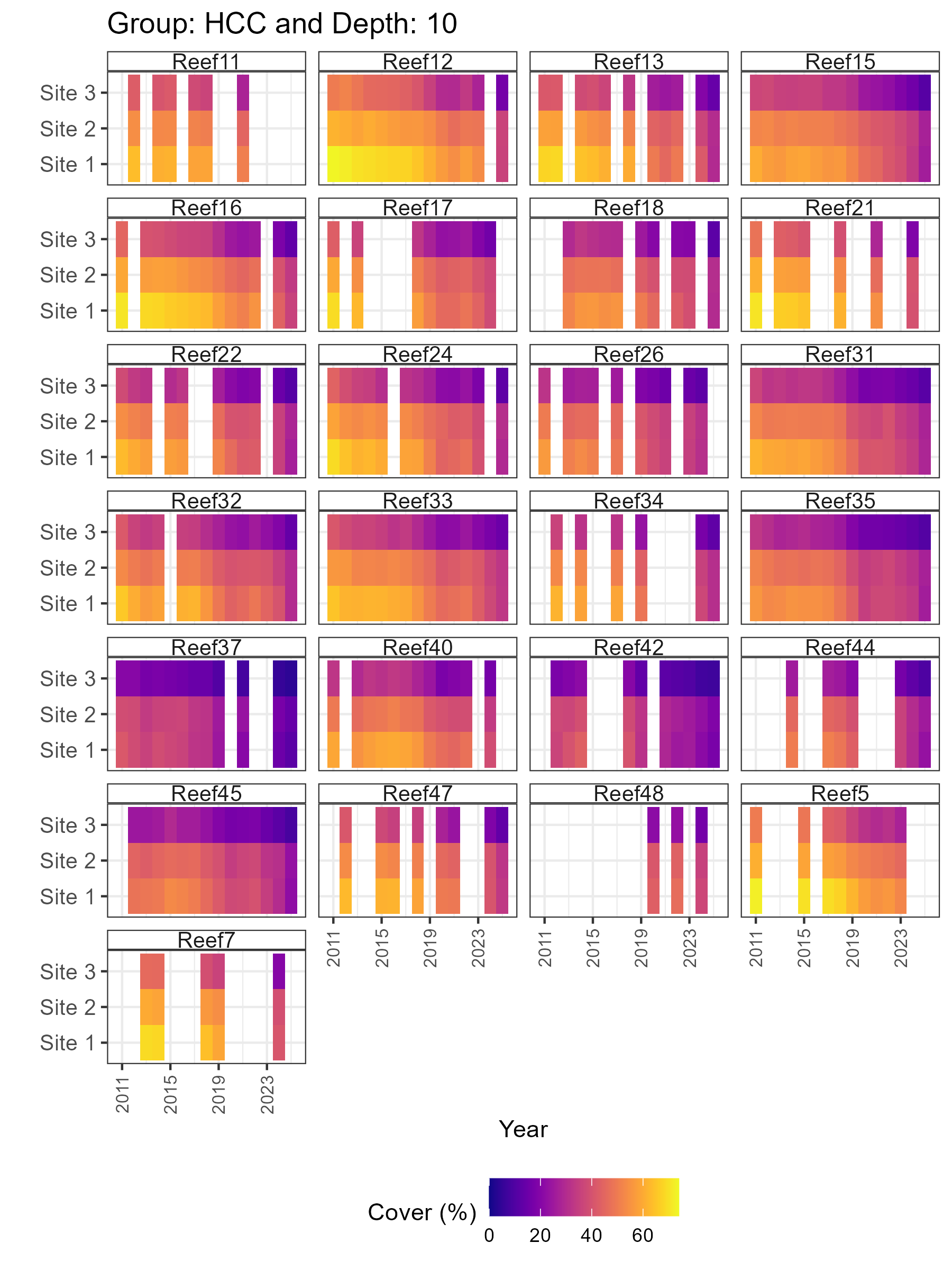
# 3.2 Heatmaps
plots_2 <- synthos::plot_synthos(synthos_data, type = "heatmaps")
purrr::walk(seq_along(plots), ~ ggsave(filename = paste0("figures/figure2.", .x, ".png"),
plot = plots[[.x]], width = 6, height = 8, dpi = 300))