Generate baselines
generate_baselines.RmdIn synthos, baselines represent the ecological community
states in the year prior to sampling. In the current implementation,
separate deterministic baseline surfaces are generated for hard coral
(HCC) and soft coral (SC). Both baselines impose a west–east spatial
gradient, with higher hard and soft coral cover in eastern locations,
reflecting broad ecological patterns encoded through simple
deterministic functions. The HCC and SC baselines differ primarily in
how latitude influences cover. For hard corals (HCC), baseline values
increase with latitude, whereas for soft corals (SC), a decreasing
latitudinal effect is imposed. In addition, the rescaled baseline ranges
are set higher for HCC than for SC, reflecting their comparatively
greater expected cover in the initial state.
1. Generate settings
surveys <- "random" # or "fixed"
data_type <- "points" # or "cover"
synthos::generateSettings(nreefs = 25, nsites = 3, nyears = 15, dhw_eff = 0.5, cyc_eff = 0.3, other_eff = 0.2)2. Generate the baseline for hard corals
## ---- SyntheticData_Spatial.mesh
mesh <- synthos::create_spde_mesh(spatial_grid,config_sp)
## Synthetic Baselines ----------------------------------------------------
spatial.grid.pts.df <- spatial_grid %>%
st_coordinates() %>%
as.data.frame() %>%
dplyr::rename(Longitude = X, Latitude = Y) %>%
arrange(Longitude, Latitude)
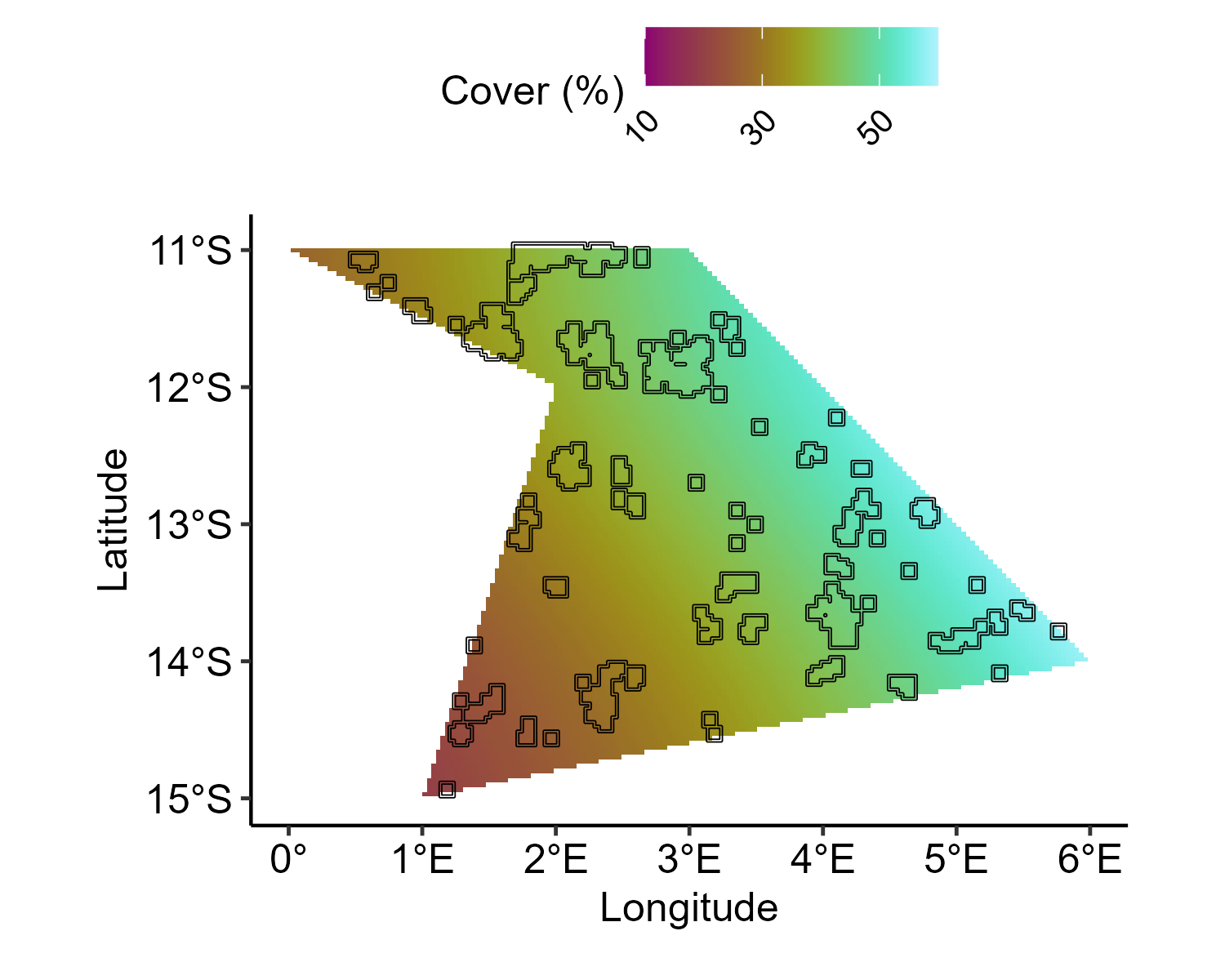
## ---- SyntheticData_BaselineSpatial.HCC
cover_range <- config_sp$hcc_cover_range
cover_range <- qlogis(cover_range)
baseline.sample.hcc <- mesh$loc[,1:2] %>%
as.data.frame() %>%
dplyr::select(Longitude = V1, Latitude = V2) %>%
mutate(clong = as.vector(scale(Longitude, scale = FALSE)),
clat = as.vector(scale(Latitude, scale = FALSE)),
Y = clong + sin(clat) +
1.5*clong + clat) %>%
mutate(Y = scales::rescale(Y, to = cover_range))
baseline.effects.hcc <- baseline.sample.hcc %>%
dplyr::select(Y) %>%
as.matrix
baseline.pts.sample.hcc <- inla.mesh.project(mesh,
loc=as.matrix(spatial.grid.pts.df [,1:2]),
baseline.effects.hcc)
baseline.pts.effects.hcc = baseline.pts.sample.hcc %>%
cbind() %>%
as.matrix() %>%
as.data.frame() %>%
cbind(spatial.grid.pts.df ) %>%
pivot_longer(cols = c(-Longitude,-Latitude),
names_to = c('Year'),
names_pattern = 'sample:(.*)',
values_to = 'Value') %>%
mutate(Year = as.numeric(Year))
ggplot() +
geom_tile(
data = baseline.pts.effects.hcc,
aes(y = Latitude, x = Longitude, fill = 100 * plogis(Value))
) +
geom_sf(
data = reefs.sf$simulated_reefs_sf,
fill = NA,
color = "black"
) +
scico::scale_fill_scico(
"Coral cover (%)",
palette = "hawaii",
direction = 1,
limits = c(10, 60),
breaks = seq(10, 60, by = 20),
labels = scales::number_format(accuracy = 1)
) +
coord_sf(crs = 4326) +
xlab("Longitude") +
ylab("Latitude") +
theme_pubr() +
theme(
legend.position = "top",
legend.justification = c(0.5, 1),
legend.direction = "horizontal",
legend.text = element_text(angle = 45, hjust = 1)
)
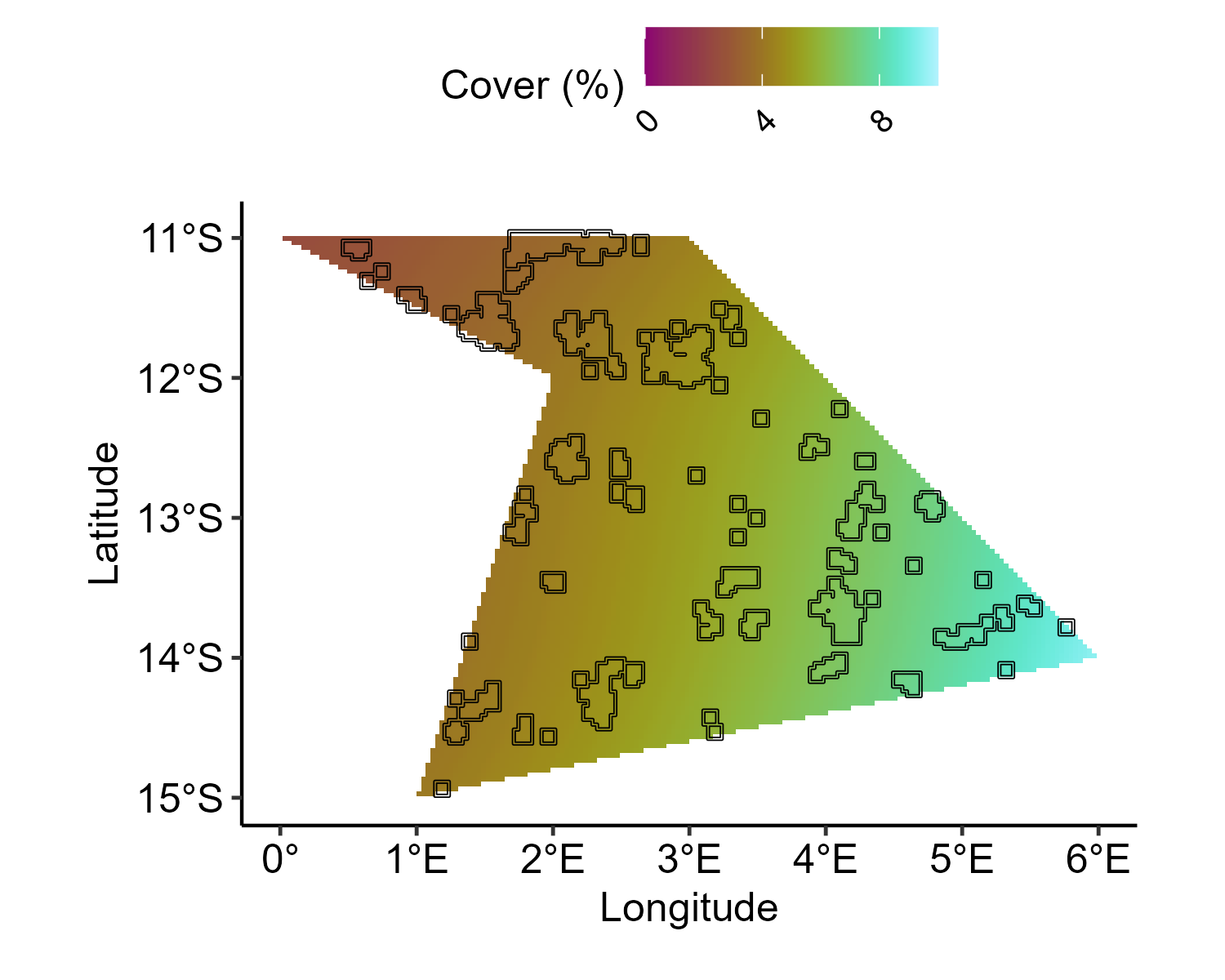
3. Generate the baseline for soft corals
cover_range <- config_sp$sc_cover_range
cover_range <- qlogis(cover_range)
baseline.sample.sc <- mesh$loc[, 1:2] %>%
as.data.frame() %>%
dplyr::select(Longitude = V1, Latitude = V2) %>%
mutate(
clong = as.vector(scale(Longitude, scale = FALSE)),
clat = as.vector(scale(Latitude, scale = FALSE)),
Y = clong + sin(clat) +
1.5 * clong + -1.5 * clat
) %>%
mutate(Y = scales::rescale(Y, to = cover_range))
baseline.effects.sc <- baseline.sample.sc %>%
dplyr::select(Y) %>%
as.matrix()
baseline.pts.sample.sc <- inla.mesh.project(mesh,
loc = as.matrix(spatial.grid.pts.df [, 1:2]),
baseline.effects.sc
)
baseline.pts.effects.sc <- baseline.pts.sample.sc %>%
cbind() %>%
as.matrix() %>%
as.data.frame() %>%
cbind(spatial.grid.pts.df) %>%
pivot_longer(
cols = c(-Longitude, -Latitude),
names_to = c("Year"),
names_pattern = "sample:(.*)",
values_to = "Value"
) %>%
mutate(Year = as.numeric(Year))
ggplot() +
geom_tile(
data = baseline.pts.effects.sc,
aes(y = Latitude, x = Longitude, fill = 100 * plogis(Value))
) +
geom_sf(
data = reefs.sf$simulated_reefs_sf,
fill = NA,
color = "black"
) +
scico::scale_fill_scico(
"Cover (%)",
palette = "hawaii",
direction = 1,
limits = c(0, 10),
breaks = seq(0, 10, by = 4),
labels = scales::number_format(accuracy = 1)
) +
coord_sf(crs = 4326) +
xlab("Longitude") +
ylab("Latitude") +
theme_pubr() +
theme(
legend.position = "top",
legend.justification = c(0.5, 1),
legend.direction = "horizontal",
legend.text = element_text(angle = 45, hjust = 1)
)